Algorithm to detect isolated space points. More...
#include <PointIsolationAlg.h>
Classes | |
| struct | Configuration_t |
| Type containing all configuration parameters of the algorithm. More... | |
Public Types | |
| using | Coord_t = Coord |
| Type of coordinate. More... | |
| using | Range_t = CoordRange< Coord_t > |
Public Member Functions | |
| template<typename PointIter > | |
| std::vector< size_t > | removeIsolatedPoints (PointIter begin, PointIter end) const |
| Returns the set of points that are not isolated. More... | |
| template<typename Cont > | |
| std::vector< size_t > | removeIsolatedPoints (Cont const &points) const |
| Returns the set of points that are not isolated. More... | |
| template<typename PointIter > | |
| std::vector< size_t > | bruteRemoveIsolatedPoints (PointIter begin, PointIter end) const |
| Brute-force reference algorithm. More... | |
| template<typename PointIter > | |
| Coord | computeCellSize () const |
Static Public Member Functions | |
| static Coord_t | maximumOptimalCellSize (Coord_t radius) |
| Returns the maximum optimal cell size when using a isolation radius. More... | |
Private Types | |
| using | Indexer_t = ::util::GridContainer3DIndices |
| type managing cell indices More... | |
| using | NeighAddresses_t = std::vector< Indexer_t::CellIndexOffset_t > |
| type of neighbourhood cell offsets More... | |
| template<typename PointIter > | |
| using | Partition_t = SpacePartition< PointIter > |
| template<typename PointIter > | |
| using | Point_t = decltype(*PointIter()) |
Private Member Functions | |
| template<typename PointIter = std::array<double, 3> const*> | |
| Coord_t | computeCellSize () const |
| Computes the cell size to be used. More... | |
| NeighAddresses_t | buildNeighborhood (Indexer_t const &indexer, unsigned int neighExtent) const |
| Returns a list of cell offsets for the neighbourhood of given radius. More... | |
| template<typename PointIter > | |
| bool | isPointIsolatedFrom (Point_t< PointIter > const &point, typename Partition_t< PointIter >::Cell_t const &otherPoints) const |
| Returns whether a point is isolated with respect to all the others. More... | |
| template<typename PointIter > | |
| bool | isPointIsolatedWithinNeighborhood (Partition_t< PointIter > const &partition, Indexer_t::CellIndex_t cellIndex, Point_t< PointIter > const &point, NeighAddresses_t const &neighList) const |
| Returns whether a point is isolated in the specified neighbourhood. More... | |
| template<typename Point > | |
| bool | closeEnough (Point const &A, Point const &B) const |
| Returns whether A and B are close enough to be considered non-isolated. More... | |
Static Private Member Functions | |
| static std::string | rangeString (Coord_t from, Coord_t to) |
Helper function. Returns a string "(\<from\> to \<to\>)" More... | |
| static std::string | rangeString (Range_t range) |
Helper function. Returns a string "(\<from\> to \<to\>)" More... | |
Private Attributes | |
| Configuration_t | config |
| all configuration data More... | |
Configuration | |
| PointIsolationAlg (Configuration_t const &first_config) | |
| Constructor with configuration validation. More... | |
| void | reconfigure (Configuration_t const &new_config) |
| Configuration_t & | configuration () const |
| static void | validateConfiguration (Configuration_t const &config) |
Algorithm to detect isolated space points.
| Coord | type of the coordinate |
This algorithm returns a selection of the input points which are not isolated. Point 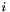 is defined as isolated if:
is defined as isolated if: 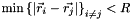 where
where 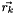 describes the position of the point
describes the position of the point 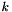 in space and
in space and 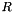 is the isolation radius.
is the isolation radius.
This class must be configured by providing a complete Configuration_t object. Configuration can be changed at any time after that.
The configuration information (Configuration_t) defines the volume the points span and the square of the isolation radius. The information on the volume may be used to optimise the algorithm, and it is not checked. If that information is wrong (that means input points lie outside that volume), the result is undefined. No check is automatically performed to assess if the configuration is valid.
The algorithm can be run on any collection of points, as long as the point class supports the PositionExtractor class. A typical cycle of use is:
The point type here is std::array<float, 3>, for which a lar::examples::PositionExtractor<std::array<float, 3>> is defined in this same library. The algorithm can be executed multiple times, and the configuration can be changed at any time (reconfigure()).
Validation of the configuration is optional, and needs to be explicitly called if desired (validateConfiguration()).
The basic method to determine the isolation of a point is by brute force, by computing the distance with all others and, as soon as one of them is found too close, declare the point non-isolated.
A refinement is implemented: the points are grouped in cubic "cells" and points in cells that are farther than isolation radius are not checked against each other. This requires some memory to allocate the structure, that can become huge. The maximum memory parameter keeps this sane.
Other refinements are not implemented. When a point is found non-isolated also the point that makes it non-isolated should also be marked so. Cell radius might be tuned to be smaller. Some of the neighbour cells may be too far and should not be checked. The grid allocates a vector for each cell, whether it's empty or not; using a sparse structure might reduce the memory; also if the grid contains pointers to vectors instead of vectors, and the grid is very sparse, there should still be some memory saving.
Definition at line 127 of file PointIsolationAlg.h.
| using lar::example::PointIsolationAlg< Coord >::Coord_t = Coord |
Type of coordinate.
Definition at line 131 of file PointIsolationAlg.h.
|
private |
type managing cell indices
Definition at line 241 of file PointIsolationAlg.h.
|
private |
type of neighbourhood cell offsets
Definition at line 244 of file PointIsolationAlg.h.
|
private |
Definition at line 247 of file PointIsolationAlg.h.
|
private |
Definition at line 250 of file PointIsolationAlg.h.
| using lar::example::PointIsolationAlg< Coord >::Range_t = CoordRange<Coord_t> |
Definition at line 132 of file PointIsolationAlg.h.
|
inline |
Constructor with configuration validation.
| first_config | configuration parameter structure |
For the configuration, see SpacePointIsolationAlg documentation. No validation is performed on the configuration.
Definition at line 155 of file PointIsolationAlg.h.
| std::vector< size_t > lar::example::PointIsolationAlg< Coord >::bruteRemoveIsolatedPoints | ( | PointIter | begin, |
| PointIter | end | ||
| ) | const |
Brute-force reference algorithm.
| PointIter | random access iterator to a point type |
| begin | iterator to the first point to be considered |
| end | iterator after the last point to be considered |
This algorithm executes the task in a  way, slow and supposedly reliable. The interface is the same as
way, slow and supposedly reliable. The interface is the same as removeIsolatedPoints. Use this only for tests.
Definition at line 575 of file PointIsolationAlg.h.
|
private |
Returns a list of cell offsets for the neighbourhood of given radius.
Definition at line 482 of file PointIsolationAlg.h.
|
private |
Returns whether A and B are close enough to be considered non-isolated.
Definition at line 606 of file PointIsolationAlg.h.
|
private |
Computes the cell size to be used.
| Coord lar::example::PointIsolationAlg< Coord >::computeCellSize | ( | ) | const |
Definition at line 439 of file PointIsolationAlg.h.
|
inline |
Returns a constant reference to the current configuration
Definition at line 168 of file PointIsolationAlg.h.
|
private |
Returns whether a point is isolated with respect to all the others.
Definition at line 522 of file PointIsolationAlg.h.
|
private |
Returns whether a point is isolated in the specified neighbourhood.
Definition at line 542 of file PointIsolationAlg.h.
|
inlinestatic |
Returns the maximum optimal cell size when using a isolation radius.
Definition at line 235 of file PointIsolationAlg.h.
|
staticprivate |
Helper function. Returns a string "(\<from\> to \<to\>)"
Definition at line 619 of file PointIsolationAlg.h.
|
inlinestaticprivate |
Helper function. Returns a string "(\<from\> to \<to\>)"
Definition at line 290 of file PointIsolationAlg.h.
|
inline |
Reconfigures the algorithm with the specified configuration (no validation is performed)
Definition at line 162 of file PointIsolationAlg.h.
| std::vector< size_t > lar::example::PointIsolationAlg< Coord >::removeIsolatedPoints | ( | PointIter | begin, |
| PointIter | end | ||
| ) | const |
Returns the set of points that are not isolated.
| PointIter | random access iterator to a point type |
| begin | iterator to the first point to be considered |
| end | iterator after the last point to be considered |
This method is the operating core of the algorithm.
The input is two iterators. The output is a collection of the indices of the elements that are not isolated. The index is equivalent to std::distance(begin, point). The order of the elements in the collection is not specified.
This method can use any collection of input data, as long as a PositionExtractor object is available for it.
Definition at line 311 of file PointIsolationAlg.h.
|
inline |
Returns the set of points that are not isolated.
| points | list of the reconstructed space points |
Definition at line 202 of file PointIsolationAlg.h.
|
static |
Throws an exception if the configuration is invalid
| std::runtime_error | if configuration is invalid |
Definition at line 407 of file PointIsolationAlg.h.
|
private |
all configuration data
Definition at line 253 of file PointIsolationAlg.h.
 1.8.5
1.8.5